
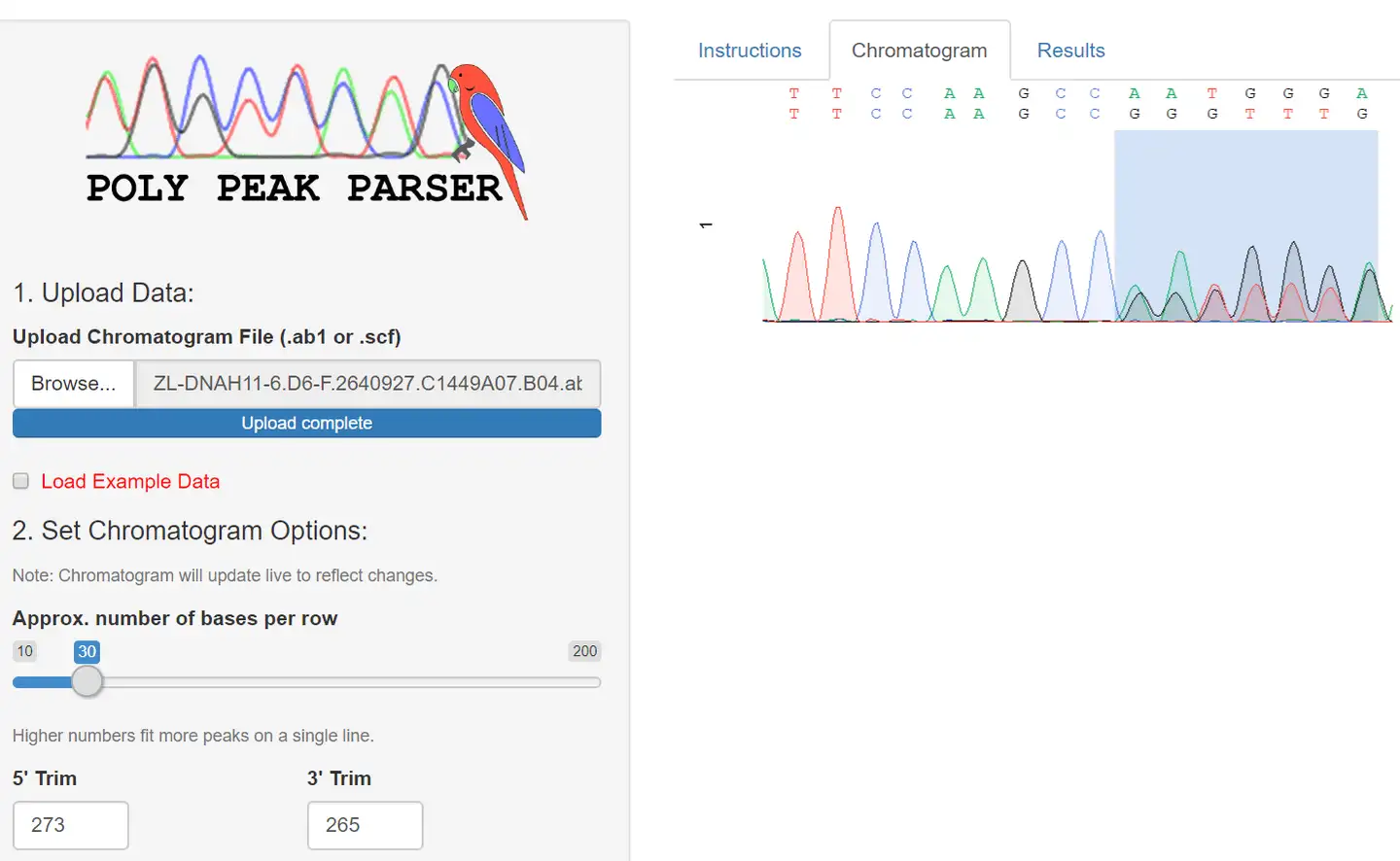
PDF) Poly Peak Parser: Method and software for identification of unknown indels using Sanger Sequencing of PCR products
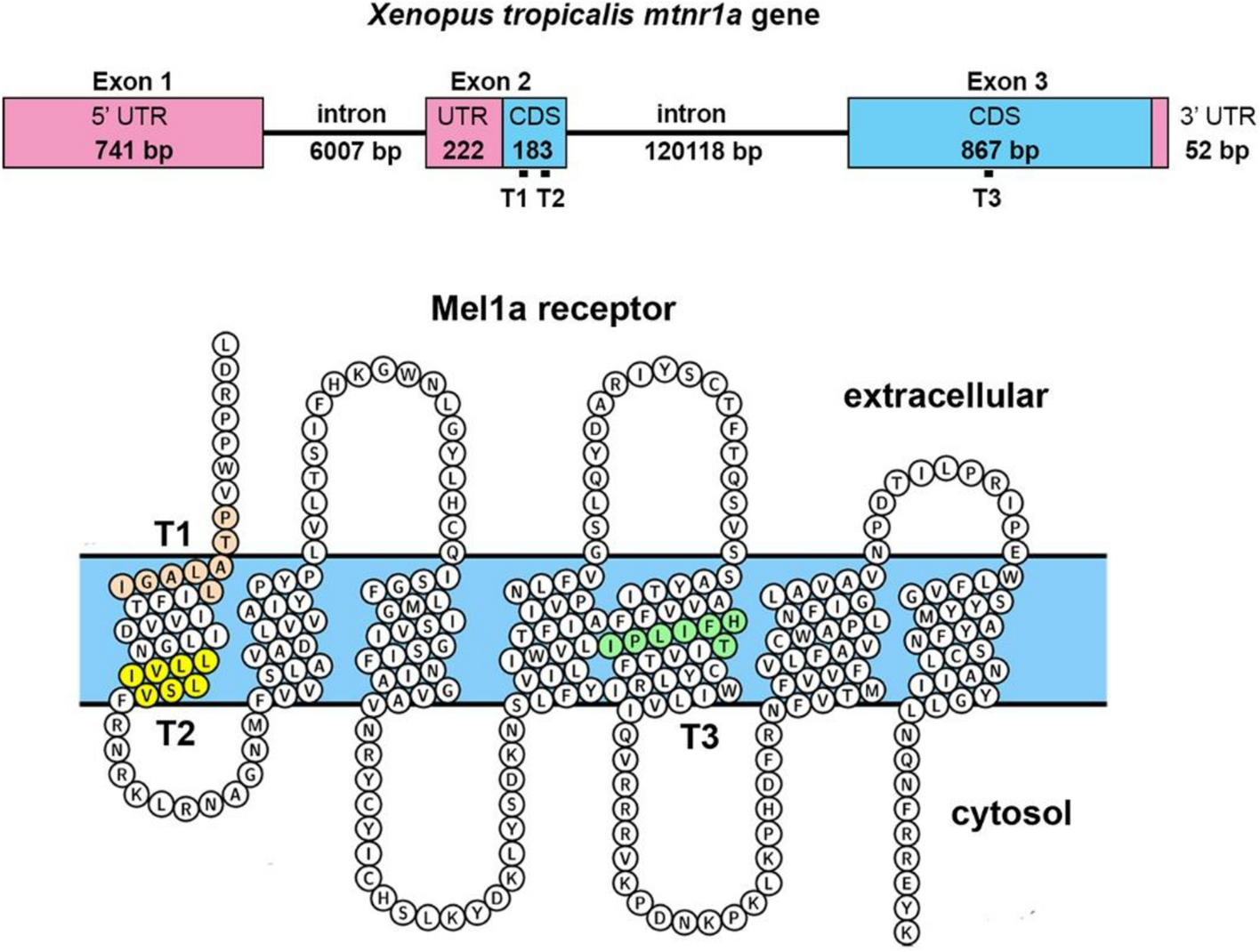
CRISPR/Cas9 mediated mutation of the mtnr1a melatonin receptor gene causes rod photoreceptor degeneration in developing Xenopus tropicalis | Scientific Reports
mRNA processing in mutant zebrafish lines generated by chemical and CRISPR-mediated mutagenesis produces unexpected transcripts that escape nonsense-mediated decay | PLOS Genetics

Tracy web applications. The upper panel shows the trace viewer employed... | Download Scientific Diagram

MultiEditR: The first tool for the detection and quantification of RNA editing from Sanger sequencing demonstrates comparable fidelity to RNA-seq: Molecular Therapy - Nucleic Acids

Testing the causality between CYP9M10 and pyrethroid resistance using the TALEN and CRISPR/Cas9 technologies | Scientific Reports

Poly peak parser: Method and software for identification of unknown indels using sanger sequencing of polymerase chain reaction products - Hill - 2014 - Developmental Dynamics - Wiley Online Library

Poly peak parser: Method and software for identification of unknown indels using sanger sequencing of polymerase chain reaction products - Hill - 2014 - Developmental Dynamics - Wiley Online Library
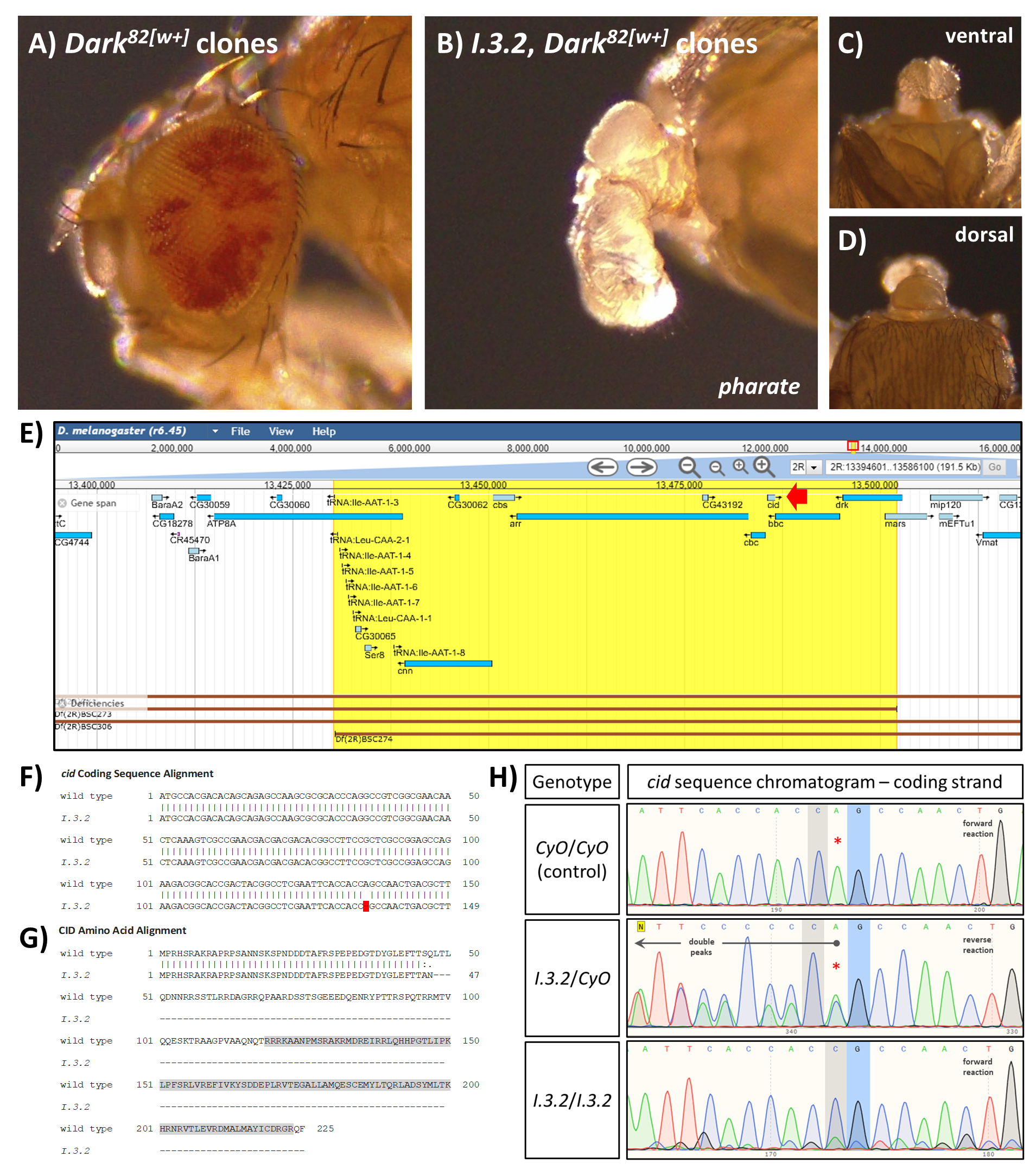
The I.3.2 developmental mutant has a single nucleotide deletion in the gene centromere identifier | microPublication

MultiEditR: The first tool for the detection and quantification of RNA editing from Sanger sequencing demonstrates comparable fidelity to RNA-seq: Molecular Therapy - Nucleic Acids
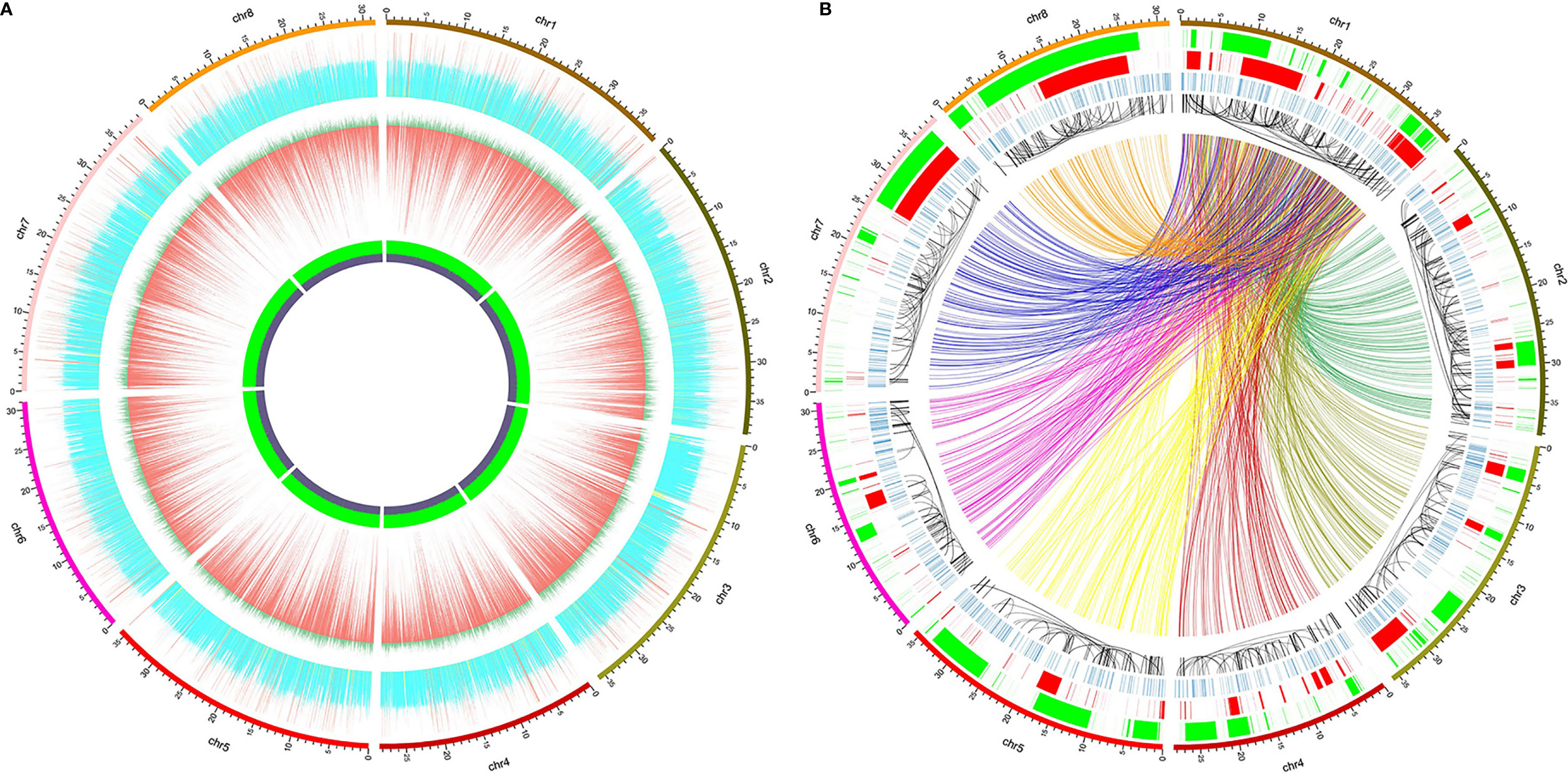
Frontiers | Genetic Diversity, Pedigree Relationships, and A Haplotype-Based DNA Fingerprinting System of Red Bayberry Cultivars

Analyses of rare predisposing variants of lung cancer in 6,004 whole genomes in Chinese - ScienceDirect
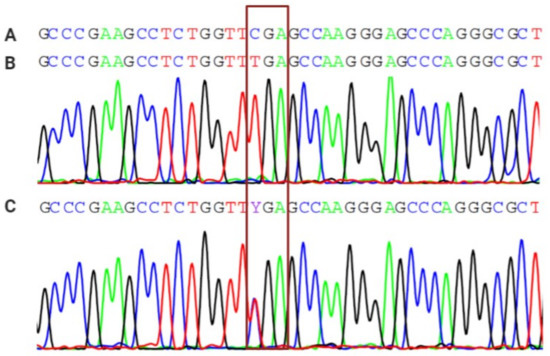



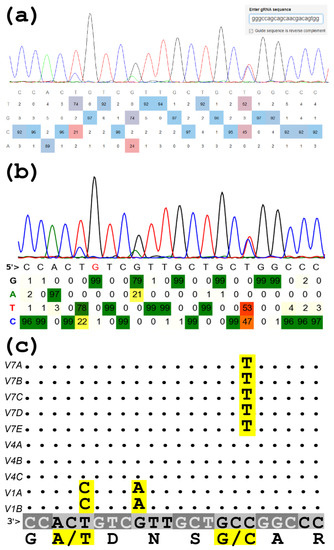
![PDF] EditR: A Method to Quantify Base Editing from Sanger Sequencing | Semantic Scholar PDF] EditR: A Method to Quantify Base Editing from Sanger Sequencing | Semantic Scholar](https://d3i71xaburhd42.cloudfront.net/c5d79e3e2af21940033a607c9951e47cff89df98/7-Figure3-1.png)



![A Python script to merge Sanger sequences [PeerJ] A Python script to merge Sanger sequences [PeerJ]](https://dfzljdn9uc3pi.cloudfront.net/2021/11354/1/fig-3-2x.jpg)
